Where you'll apply the knowledge and techniques you've acquired throughout the previous modules.
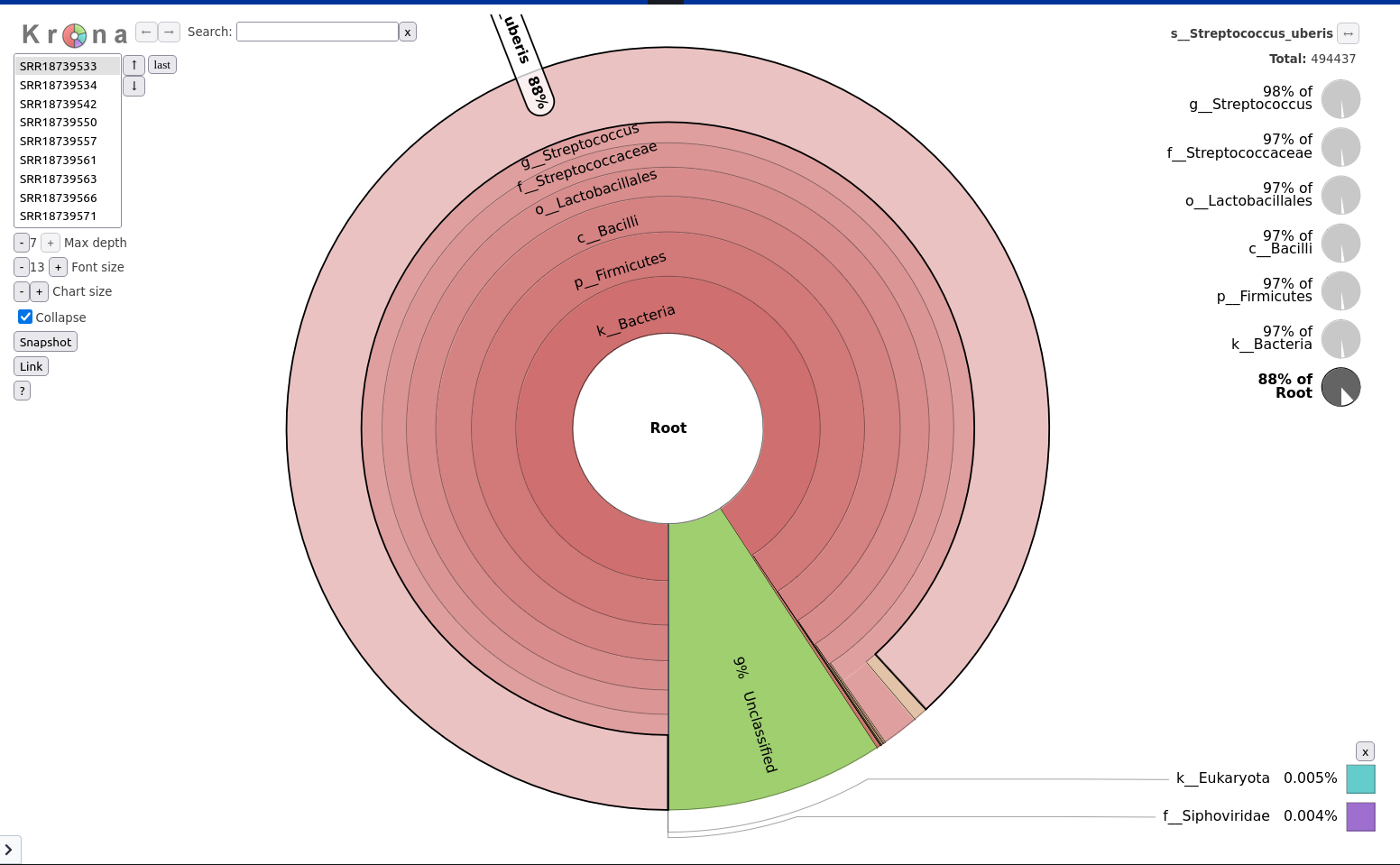
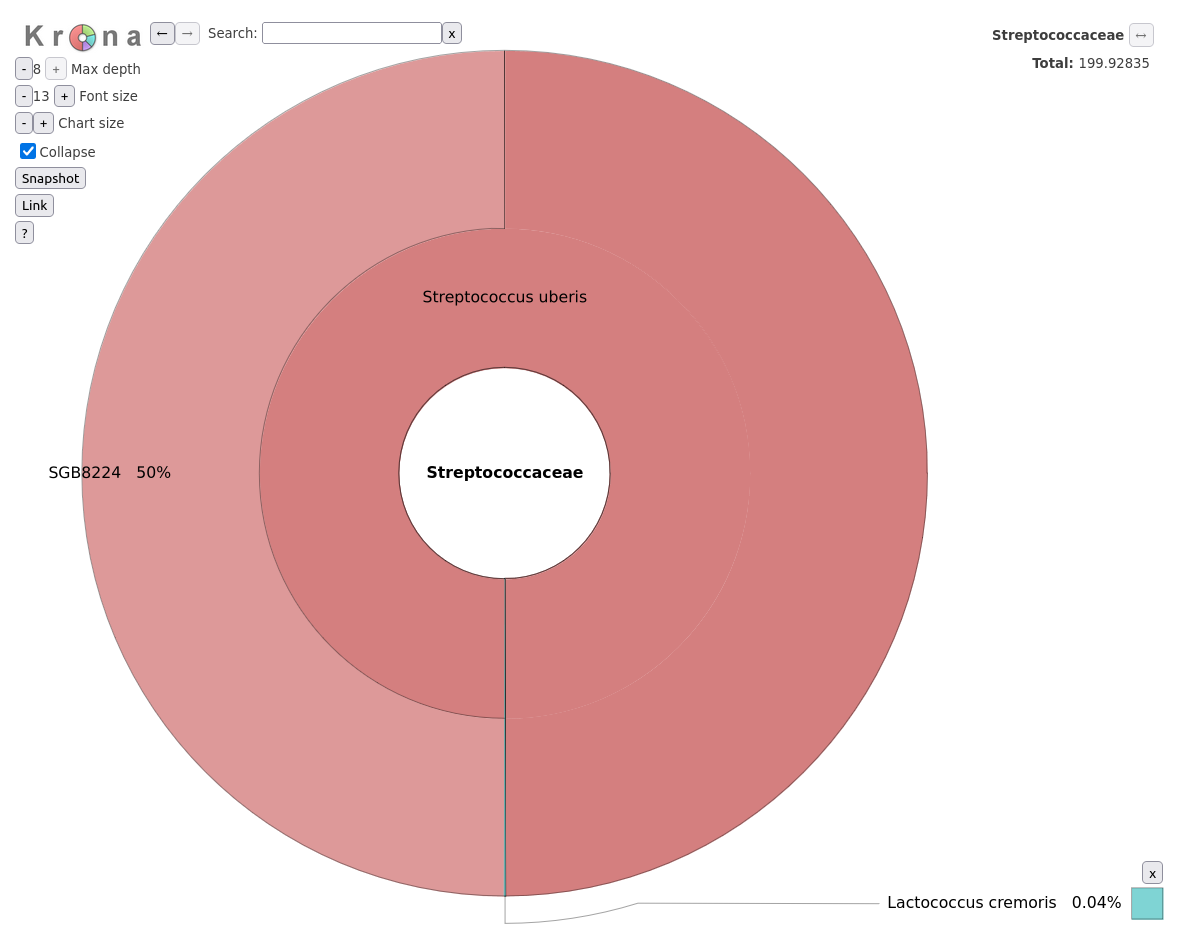
In this engaging “Hands-On” section, we embark on an exciting journey to replicate the genomic analysis performed in the study titled Genomic Surveillance Reveals Antibiotic Resistance Gene Transmission via Phage Recombinases within Sheep Mastitis-Associated Streptococcus uberis.
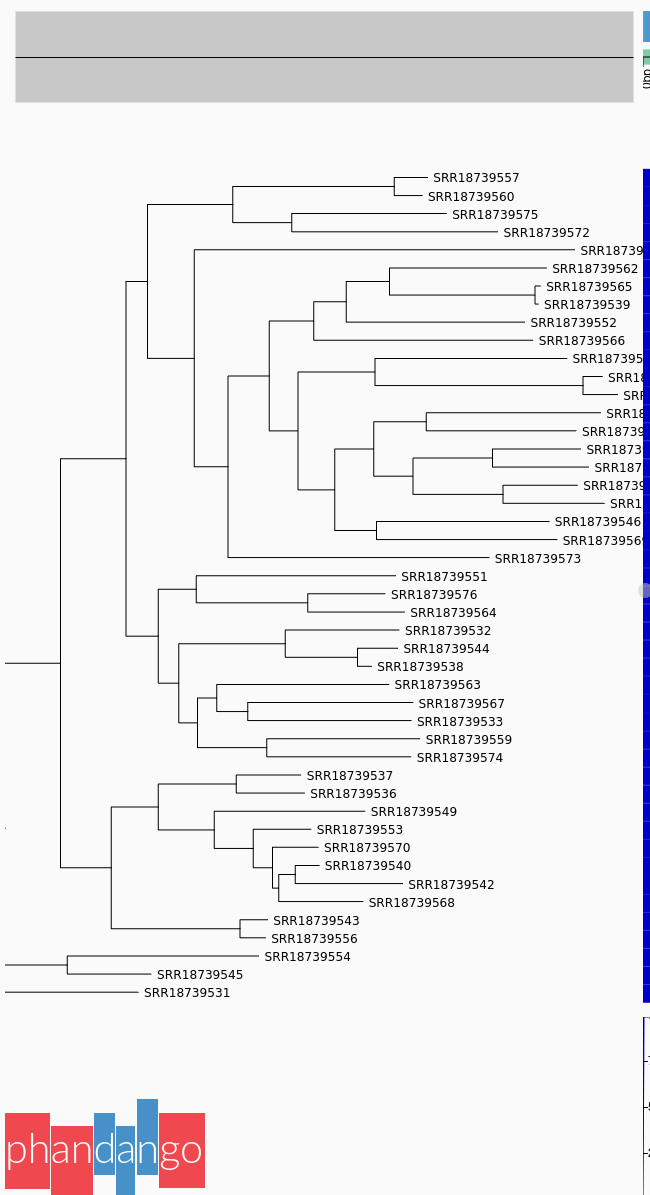
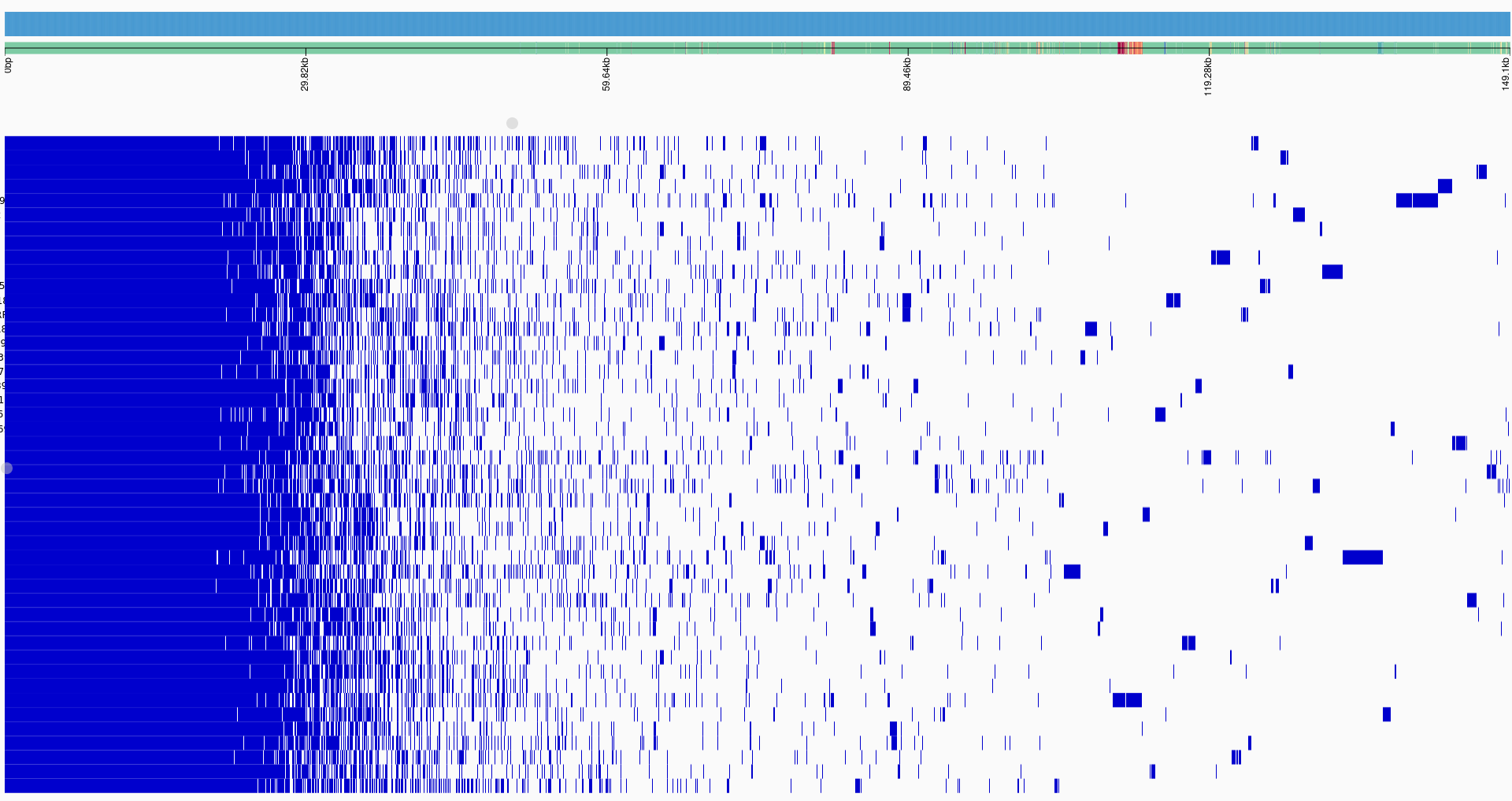
This study, conducted on 46 S. uberis isolates collected from mastitis-infected sheep in Sardinia, Italy, serves as our real-world case study.
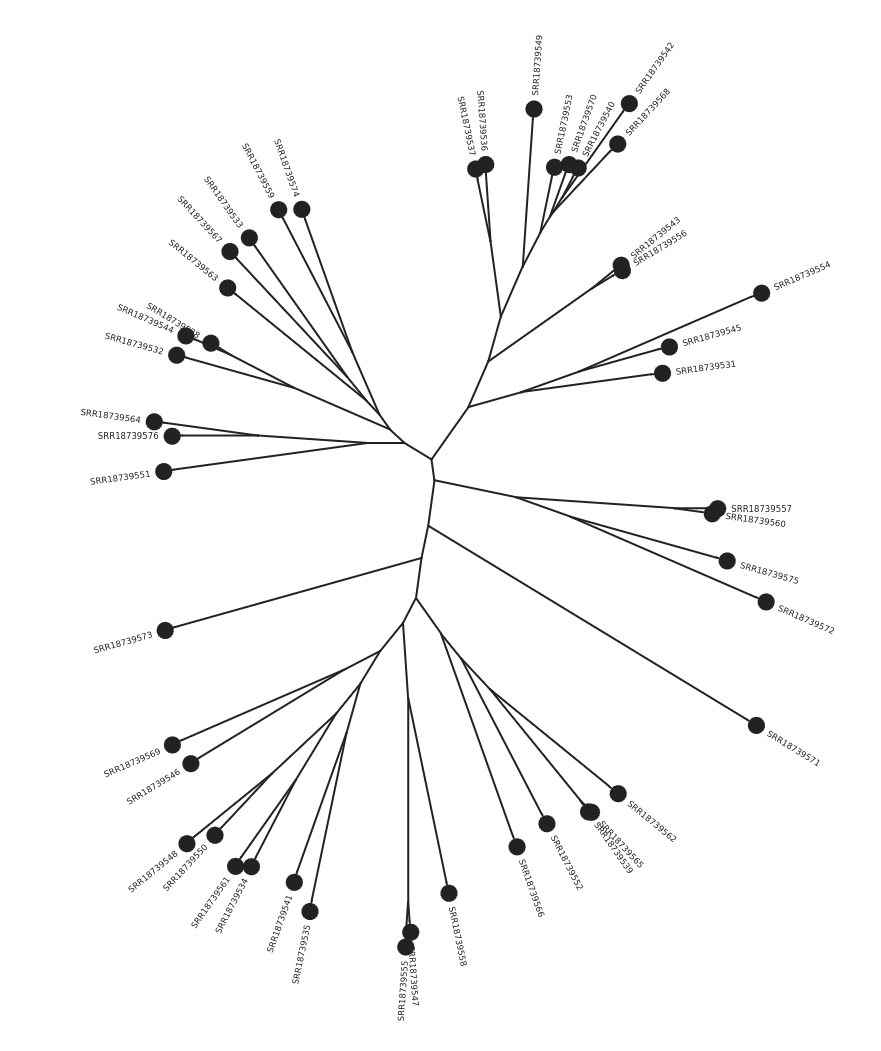
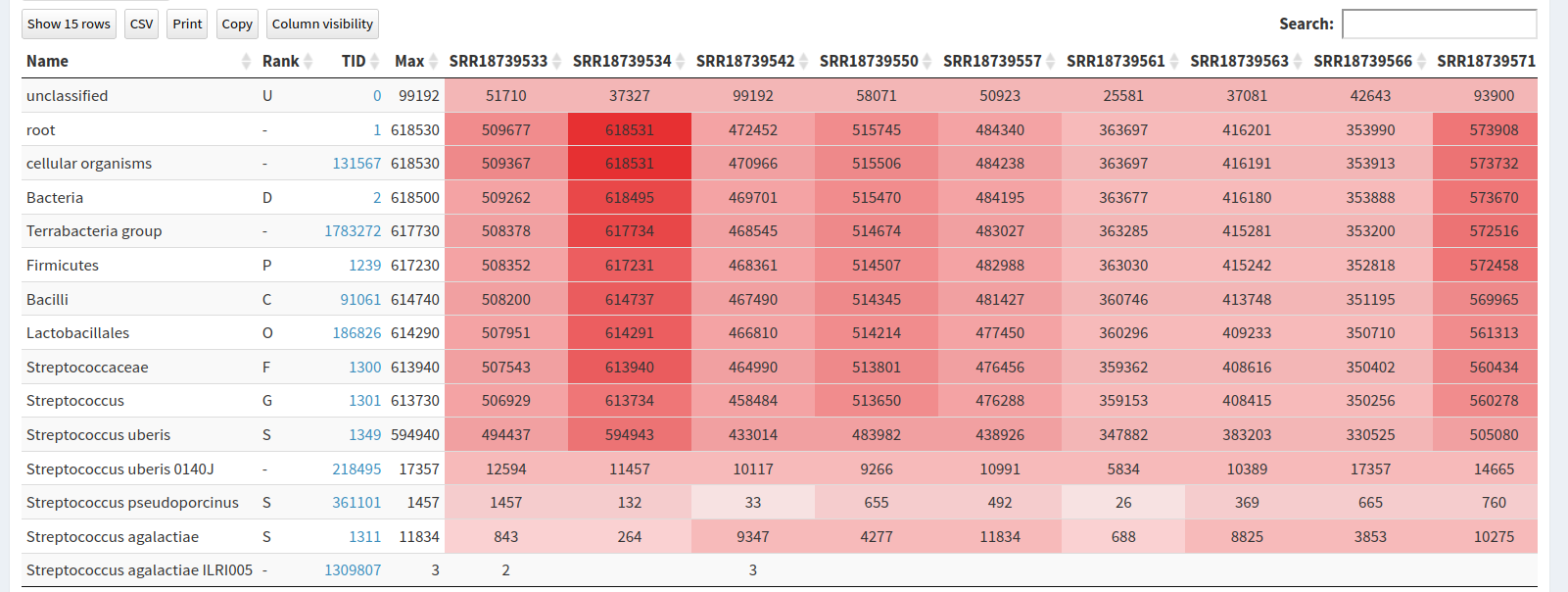
PopPUNK, which found 27 distinct isolate
clusters, indicating considerable genetic variability consistent with environmental isolates. Geographic
trends were identified including regional linkage of several isolate clusters. Multi-locus Sequence Typing
(MLST) performed poorly and provided no new insights.Retrieve the BioProject ID from the article and upload the datasets to a new history in Galaxy
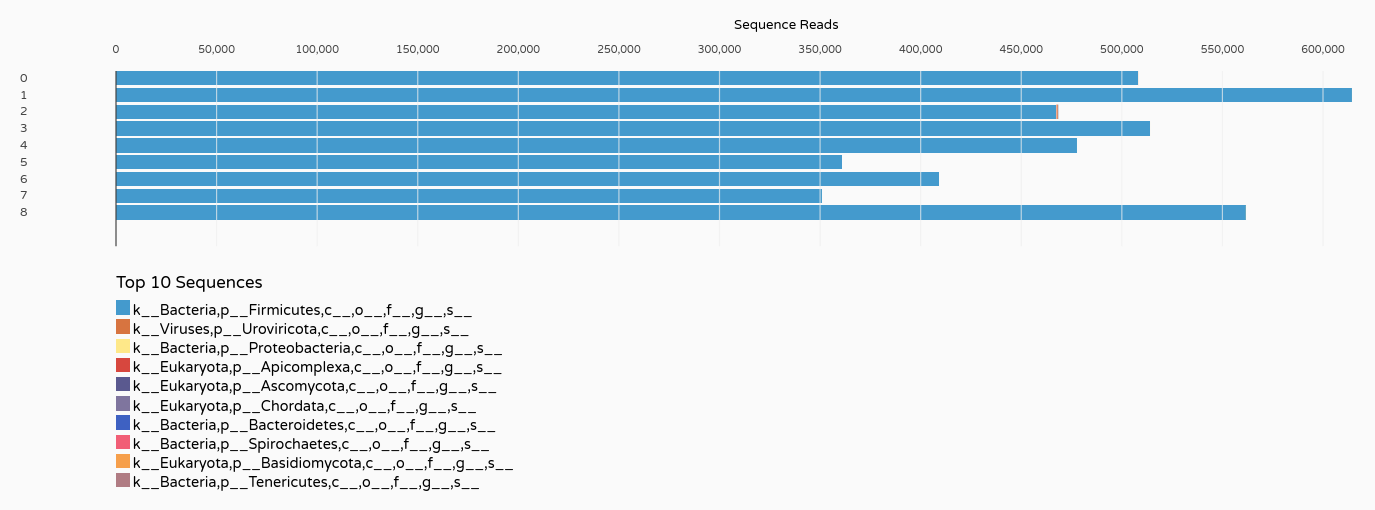
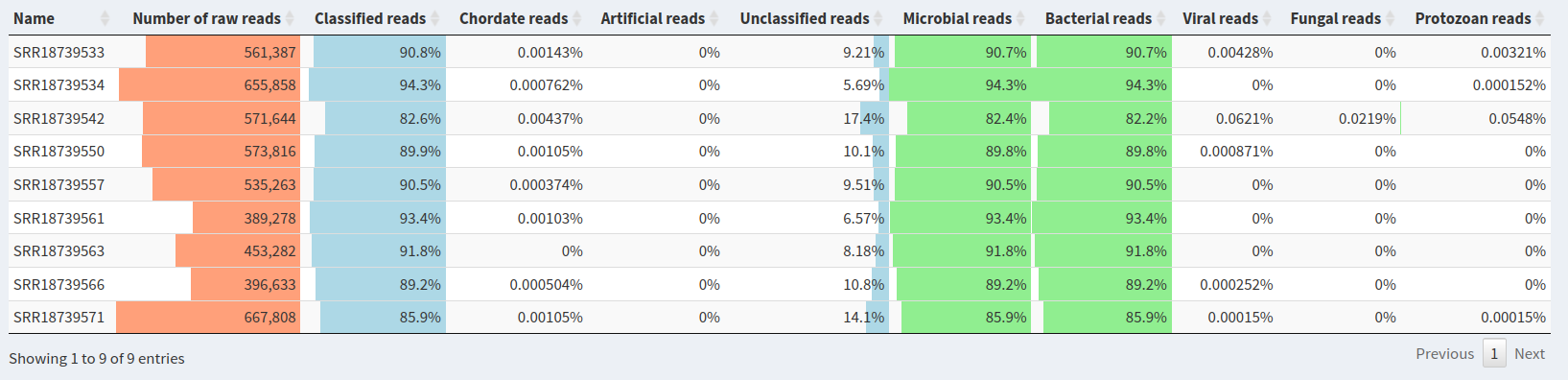
In order to save time, we have pre-selected the samples that appeared to be the most significant.
SRR18739533
SRR18739534
SRR18739542
SRR18739550
SRR18739557
SRR18739561
SRR18739563
SRR18739566
SRR18739571
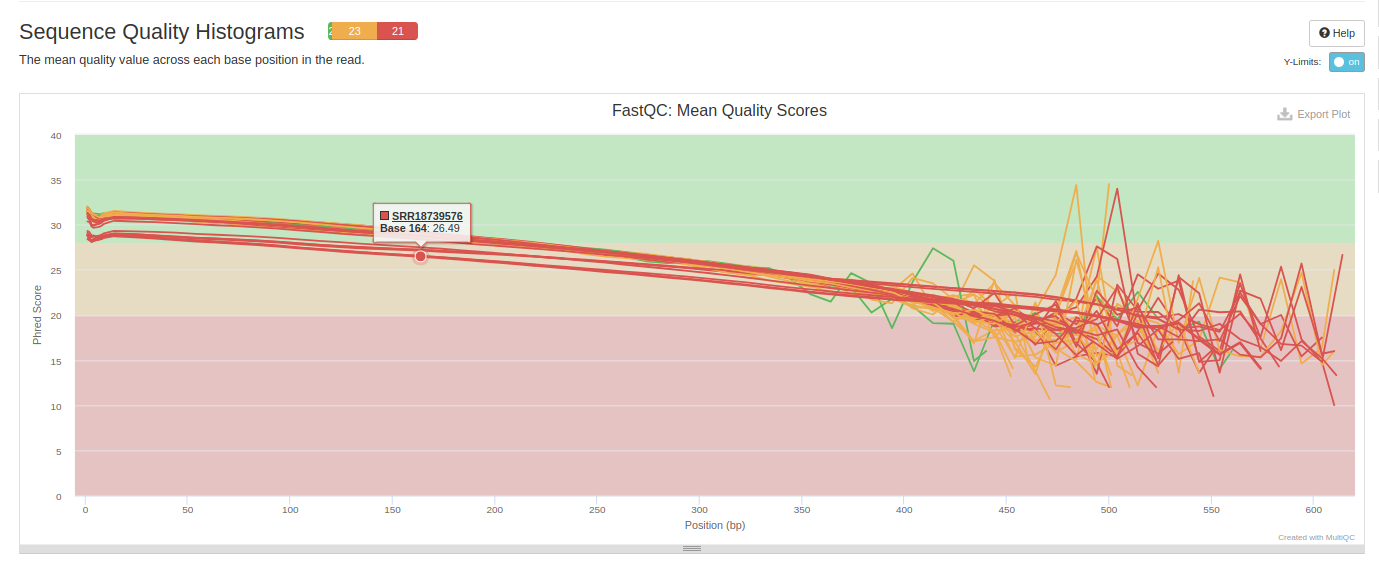
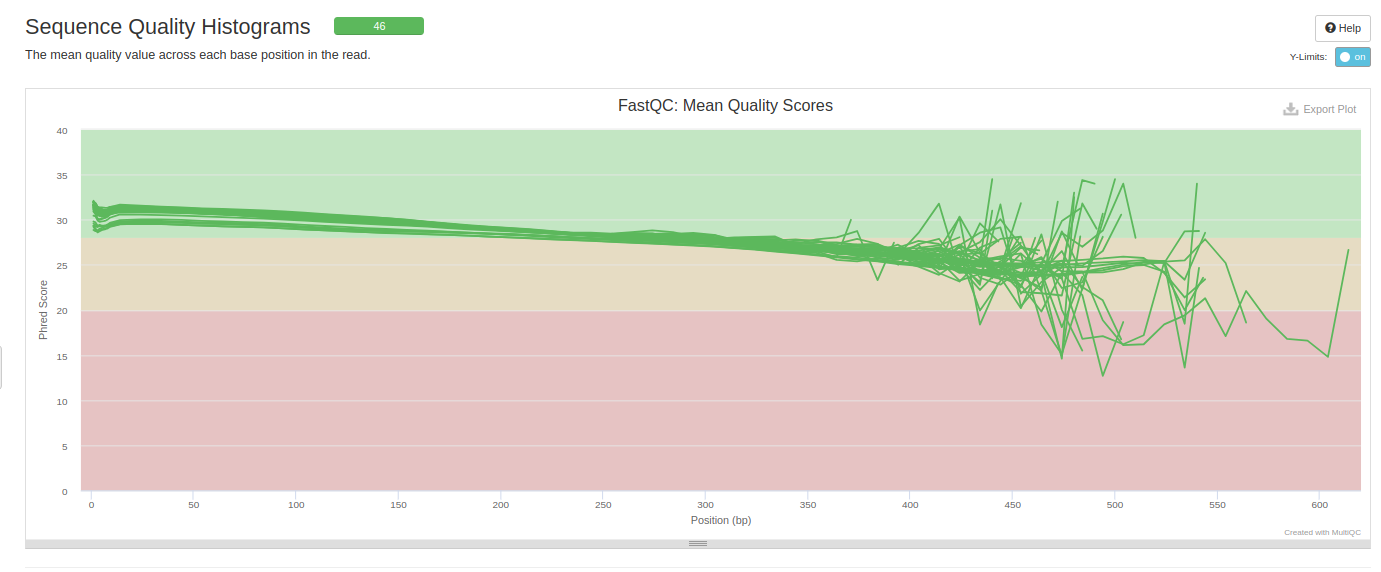
NOTE!!!
metaSPAdes only accepts paired-end reads.MEGAHIT accepts single reads but for single reads can be used in individual mode for several samples.SPAdes provides Ion Torrent featuresUnicycler can be used in individual mode for several samples, even if it is not expressly developed for ion torrent reads
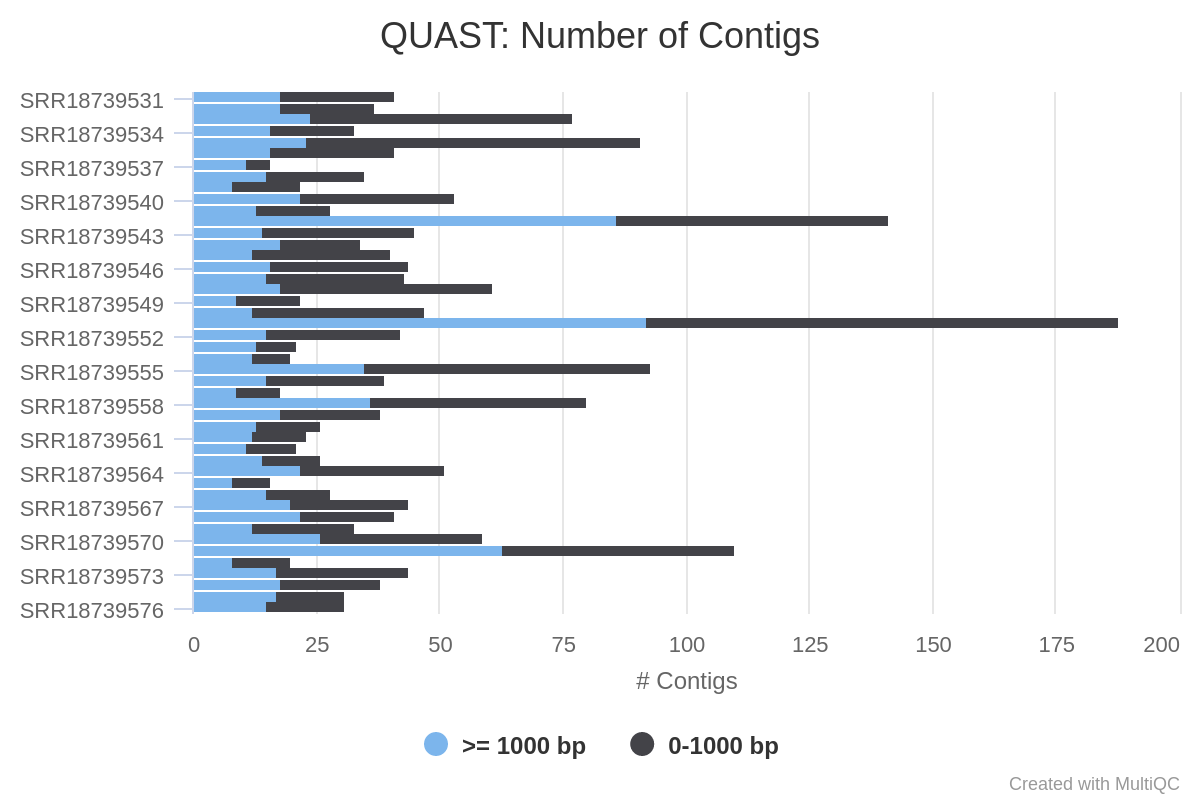
SPAdes
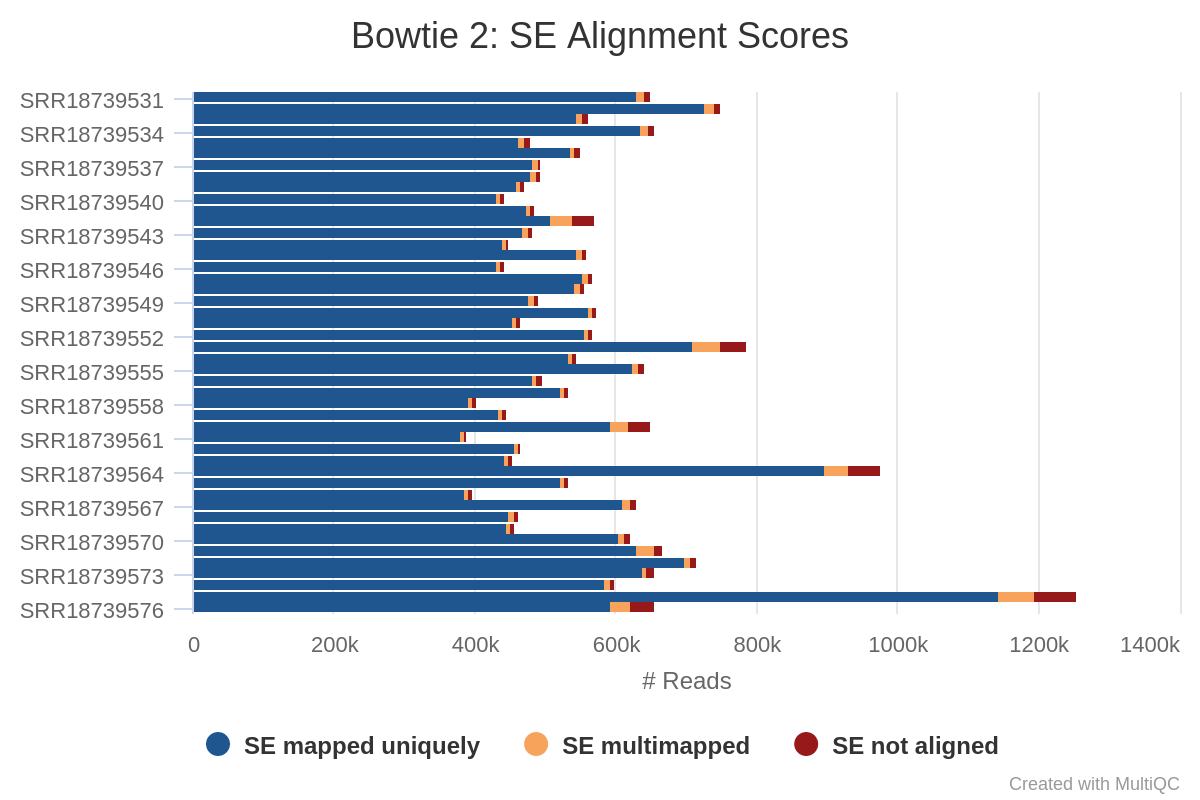
Unicycler