Pangenome Analysis
If we analyse DNA from several bacterial strains, we may want to know which genes they have in common and which are unique to some strains.
Pangenomic analysis studies the diversity and evolution of genes and genomes within a group of related organisms.
It goes beyond traditional single-genome analysis by examining the collective genomic content of a population,
species, or even a larger taxonomic group.
- The core genome is the group of genes shared by every genome in the tested set. Gene sequences are similar but not necessarily identical. Core genome SNPs are those SNPs found in the genes in the core genome; i.e. at a particular site, the nucleotide varies. We can use these SNPs to infer relationships between the strains. Some authors have divided the core pangenome in
- hard core: families of homologous genes that has at least one copy of the family shared by every genome (100% of genomes)
- soft core: those families distributed above a certain threshold (90%)
- The accessory genome is the group of genes that are not in all the strains.
- Shell: is the part of the pangenome shared by the majority of the genomes in a pangenome. There is not a universally accepted threshold to define the shell genome, some authors consider a gene family as part of the shell pangenome if it shared by more than 50% of the genomes in the pangenome
- Cloud: consists of those gene families shared by a minimal subset of the genomes in the pangenome, it includes singletons or genes present in only one of the genomes. It is also known as the peripheral genome. Gene families in this category are often related to ecological adaptation
- The pan genome is the sum of the core and accessory genes. That is, a combination of all the genes that are found in the clade of interest.
In a study that involves the pangenomes of Bacillus cereus and *_Staphylococcus aureus_,
some of them isolated from the international space station, the thresholds used for segmenting the pangenomes were as follows:
- Cloud: presence in <10% of the genomes
- Shell: presence in 10% to 95% of the genomes
- Core , presence in >95% of the genomes
Recently several stand-alone or server-based suites have become available for pangenome analysis:
a Review of Pangenome Tools and Recent Studies can be found here
We decided to use Roary
a high speed stand alone pan genome pipeline,
which takes annotated assemblies in GFF3 format (produced by Prokka) and calculates the pan genome.
Also using a standard desktop PC, it can analyse datasets with thousands of samples,
something which is computationally infeasible with other existing methods,
without compromising the quality of the results.
For example: 128 samples can be analysed in under 1 hour using 1 GB of RAM and a single processor.
To perform this analysis using existing methods would take weeks and hundreds of GB of RAM.
Go Up
Preparing Data
Roary accepts the annotated genome in GFF3 format as input, per sample (e.g. output from Prokka),
with the strict requirement that the samples must belong to the same species.
For this tutorial, we will use sequences from different strains of Staphylococcus aureus:
Go Up
Download datasets
- click on link
Send toComplete RecordFile- Format: FASTA
-
Create File
-
Rename file with strain label
Go Up
Upload datasets on Galaxy
NOTE! Please, use
- Create a new history and rename it
- Upload data
- Create a collection named strains
Go Up
Annotate Genomes
Prokka Tool with the following parameters (leave everything else unchanged):
- contigs to annotate: strains
We will not be running the entire workflow during this tutorial: it can be quite time-consuming and resource-intensive.
I have prepared the output of the workflow in advance.
Go Up
Pangenomic Analysis with Roary
Roary is widely used in microbiology and bacterial genomics research to study the genomic diversity of bacterial populations,
track the evolution of pathogens, and understand the genetic basis of phenotypic traits.
Starting with a set of annotated prokaryotic genomes, in standard formats like GFF, Roary will be able to:
- converts coding sequences into protein sequences
- clustered these protein sequences by several methods
- further refines clusters into orthologous genes
- for each sample, determines if gene is present/absent: produces the file gene_presence_absence.csv
- uses this gene p/a information to build a tree, using
FastTree: produces the file accessory_binary_genes.fa.newick
- overall, calculates number of genes that are shared, and unique: produces the file summary_statistics.txt
- aligns the core genes (if option used, as above) for downstream analyses
Roary Tool with the following parameters:
- Individual gff files or a dataset collection: Collection
- Dataset collection to submit to Roary: GFF collection from
Prokka
There are three output files.
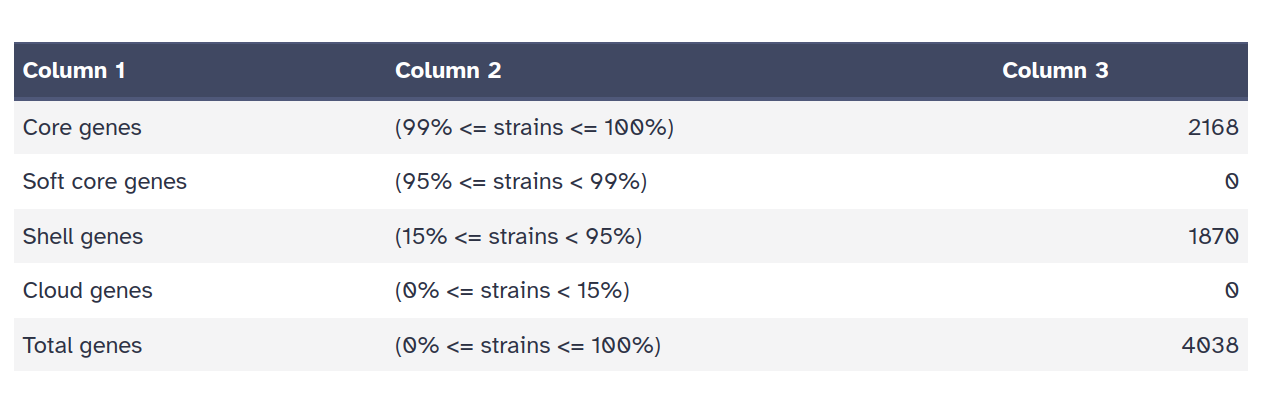
- Summary statistics: a table of counts of shared genes (core genome) and total genes (pan genome).
- Core gene alignment: FASTA file with core genes aligned
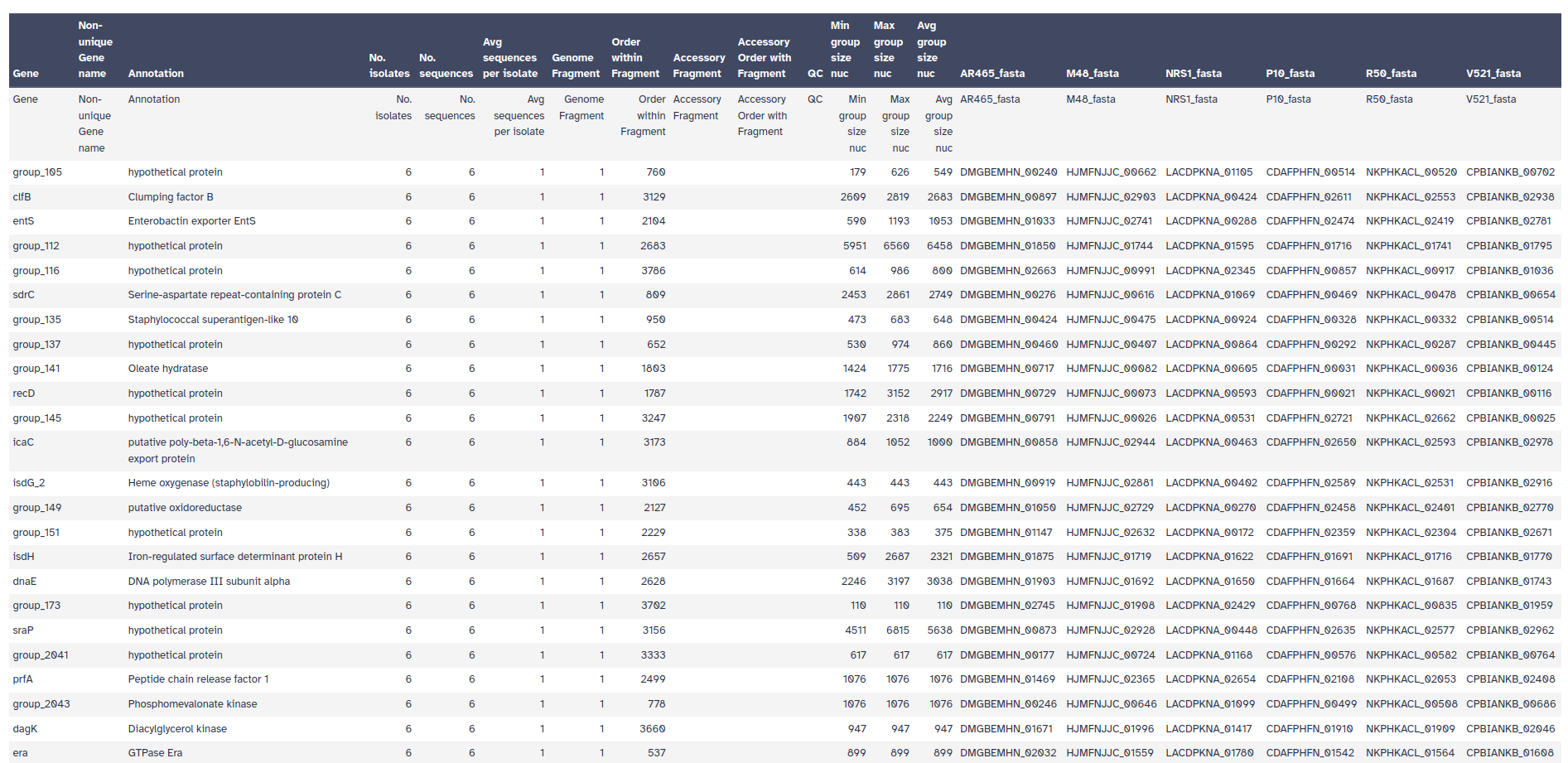
- Gene presence/absence: for each sample, determines if gene is present/absent
- Column 3 shows the annotated gene name.
- Column 4 shows the number of isolates that the gene was found it
Go Up
Infer phylogeny using core gene snps
Roary has produced an alignment of the core genes. We can use this alignment to infer a phylogenetic tree of the isolates.
Phylogeneitc reconstruction with RaXML Tool with the following parameters:
- Source file with aligned sequences: Core gene alignment from
Roary
- Model Type: Nucleotide
- Substitution Model: GTRGAMMA
- Inspect the six output files
-
Click on Result.
-
Under the file, click on the Visualize icon (a graph)
- Choose
PhyloViz
- These isolates are all very closely related and so the structure of the tree is narrow.
- To expand, go to the right hand box for *Phyloviz Settings.
- Change the Phylogenetic Spacing to 2500 and the Vertical Spacing to 30.

- Choose
Phylogenetic Tree Visualization
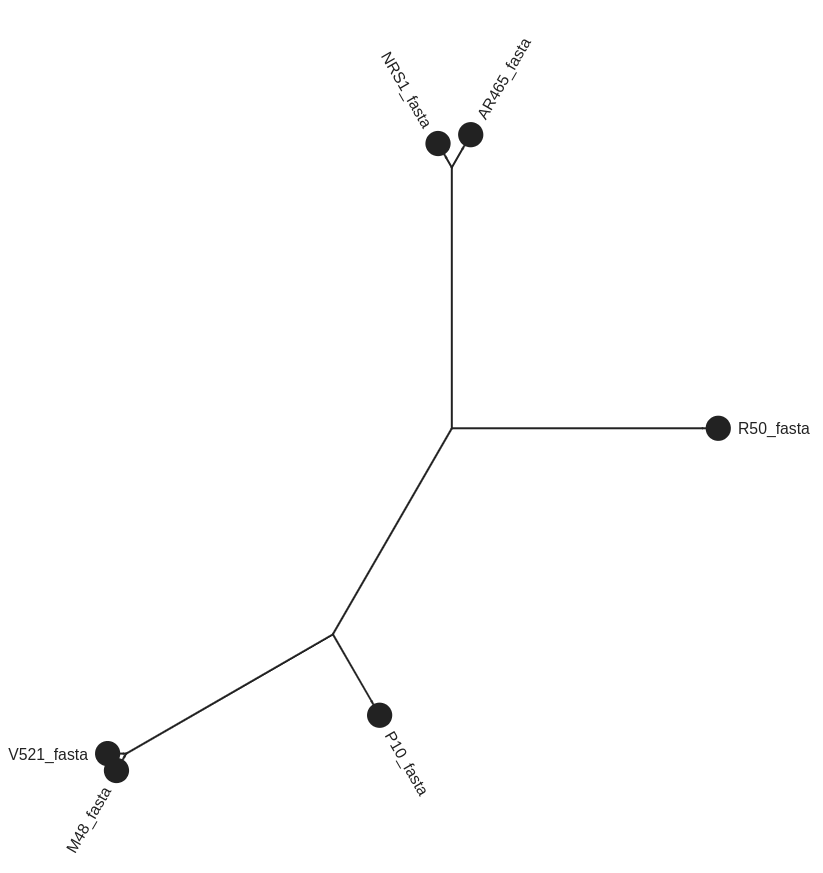
Go Up
Vizualize with Phandango
- Download Result from
Phylogeneitc reconstruction with RaXML
- Rename as raxml.tree
- Download Gene presence/absence from
Roary
- Rename as gene_presence_absence.csv
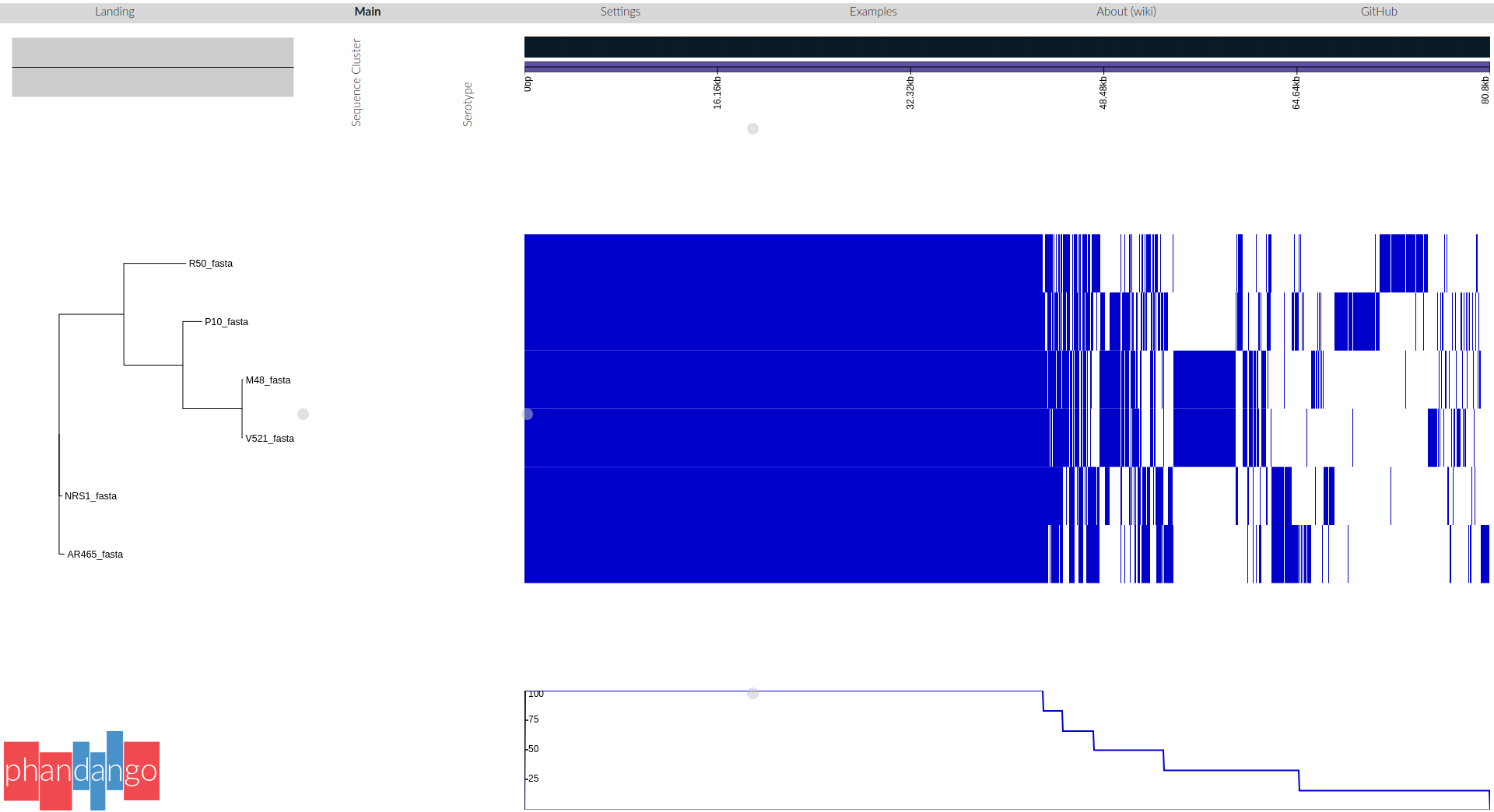
- Go to http://phandango.net/
- Drag and drop the two files onto the landing page.
- view the tree of samples and their core and pan genomes
- each blue coloured column is a gene: genes are present or absent in each isolate
- the core genes are shared by all isolates
Go Up
Hands-On
Now, it’s your turn!
Go Up








